1 Introduction
Citizen science games have, in recent years, begun to actively engage the general public and participants from a variety of backgrounds in scientific research [Cooper et al., 2010 ]. In these projects, game elements [Deterding et al., 2011 ] may be used for motivating players to participate in tasks that are difficult to automate and might otherwise be tedious or lack motivating rewards. Several such projects involve participants in manipulating complex 3D structures. Eyewire [Kim et al., 2014 ], for example, involves players in reconstructing the 3D shape of neurons, and Foldit [Cooper et al., 2010 ] involves players in 3D protein structure manipulation. However, these spatial and structural manipulations are typically carried out using the mouse, which allows only one point of interaction. Touchscreens with multi-touch support can allow more flexible and powerful interactions with 3D structures. Could citizen science game projects benefit from the proliferation of these devices and screens by implementing multi-touch manipulations?
This work, in particular, looks at the 3D protein manipulation interface of Foldit [Cooper et al., 2010 ]. Foldit is an online protein folding and design game, which provides players with numerous tools for interactively modifying protein structures, seeking to find a structure’s global energy minimum; that is, the configuration of the protein predicted to have the lowest energy. Foldit is built on top of the powerful Rosetta molecular modeling suite [Leaver-Fay et al., 2011 ], which provides not only the protein structure model, but also an energy function used to provide players with feedback on the quality of their structure. Foldit is targeted at citizen scientists, who come from a variety of backgrounds and familiarity levels with biochemistry [Cooper et al., 2010 ].
With the proliferation of smartphones and tablets, touch screen interfaces have become widely popular for games. Today, mobile games are played by millions of people using touch interfaces every day. In order to maximize the potential contributors to scientific discovery in Foldit we designed and implemented a touch interface for the game, based on standard interaction techniques for touch interfaces [Jankowski and Hachet, 2015 ]. We evaluated the interface in a comparative user study with 41 subjects against the standard mouse based interface. In our user study comparing both interfaces, we found that participants performed very similarly using multi-touch and mouse interaction for Foldit. However, the results indicate that for tasks involving guided rigid body manipulations (e.g. translation and rotation) the multi-touch interface may have benefits, while for tasks involving direct selection and dragging of points the mouse interface may have benefits. Even though subjects overall had no preference for one or the other interface and no differences in subjective usability, enjoyment or mental demand were present, participants reported higher attention and spatial presence using the multi-touch interface.
This work contributes an examination of mouse and multi-touch use for interactive protein manipulation with feedback from an energy function, by a mix of scientifically novice and experienced users in two types of tasks. The overall similar results for touch and mouse based interfaces indicate that Foldit can be played on touch screens without compromises in usability and player performance, potentially opening the game for a bigger player base. Further, use of multi-touch may improve outcomes where rigid body manipulations are needed, such as docking multiple protein structures.
2 Background
In this section, we give an overview of the related work for mouse and multi-touch based interfaces as well as manipulation of molecular structures.
2.1 Mouse and multi-touch
Multiple studies have explored the differences between direct touch input and mouse input. Watson et al. [Watson et al., 2013 ] studied the difference in participant performance and overall experience during game playing and concluded that direct touch input improved performance, with regard to speed and accuracy, as well as resulting in a more positive overall experience, with regard to enjoyment, motivation, positive feeling, and immersion of system use. These results are also compatible with the research done by Drucker et al. [Drucker et al., 2013 ], who studied user performance in a touch interface based on a traditional WIMP (Windows, Icons, Menus, and Pointers) or FLUID interface design. Further research also showed that direct touch input is preferential to mouse input, in terms of overall speed and subjective enjoyment. Knoedel et al. [Knoedel and Hachet, 2011 ] showed that rotations, scaling, and translations (RST) from direct-touch gestures shorten completion times, but indirect interaction improves efficiency and precision. Despite higher overall performances and enjoyment levels with touch input, it has been shown in some studies that touch is also related to less accurate cursor positioning than mouse input. This inconsistency has been attributed to potential differences in target size. Contrary to other studies, a recent comparison of mouse, touch, and tangible input for 3D positioning tasks by Besançon et al. [Besançon et al., 2017 ] showed that they are all equally well suited for precise 3D positioning tasks.
Interacting with 3D content on screens comprises special challenges covered by many publications in research. Particularly, the gap between the two-dimensional inputs and performing a three-dimensional interaction raises problems. Mouse-based applications often use transformation widgets in order to deal with this problem. Cohe et al. developed a 3D transformation widget especially designed for touchscreens [Cohé, Dècle and Hachet, 2011 ]. Developing and evaluating touch-based interactions for independent fine-grained object translation, rotation and scaling Wu et al. [Wu et al., 2015 ], could show that direct manipulation based interactions outperform widget-based techniques regarding both the efficiency and fluency. However, Fiorella et al. [Fiorella, Sanna and Lamberti, 2010 ] showed that when it comes to fine control of objects differences between direct manipulation and GUI based manipulation interfaces are strongly reduced. Making 3D rotation accessible to novice users on touchscreen was looked at by Rousset et al. [Rousset, Bérard and Ortega, 2014 ], who designed two-finger based gestures with a surjective mapping. Different dedicated gestures have been developed using two or more fingers to control 6 DOFs (degrees of freedom) for the manipulation of 3D Objects [Herrlich, Walther-Franks and Malaka, 2011 ; Liu et al., 2012 ; Reisman, Davidson and Han, 2009 ]. Hancock et al. developed different rotation and translation techniques for tabletop displays [Hancock, Carpendale, Vernier et al., 2006 ] and compared one, two, and tree touch point based rotation and translation techniques [Hancock, Carpendale and Cockburn, 2007 ]. Based on this work they also developed a force-based interaction in combination with full 6 DOF manipulation [Hancock, Ten Cate and Carpendale, 2009 ]. Today a wide variety of direct manipulation gestures are available, which are well understood in terms of performance and ergonomics [Hoggan, Nacenta et al., 2013 ; Hoggan, Williamson et al., 2013 ; Jankowski and Hachet, 2015 ].
2.2 Interactive molecular manipulation
Interactive manipulation of computational models of proteins and other molecular structures has been the subject of study for several decades, with applications in understanding naturally-occurring structures as well as designing novel synthetic ones with new functions. Early work by Levinthal [Levinthal, 1966 ] and the Sculpt system developed by Surles et al. [Surles et al., 1994 ] explored tools for manipulating proteins with aims of energy minimization.
Since then, a variety of scenarios involving novel input technologies for molecular manipulation have been explored and evaluated. These include improving performance using force feedback in a simplified 6 DOF spring-based energy minimization [Ming, Beard and Brooks, 1989 ]; bimanual scientist creation of molecular structures and animations [Waldon et al., 2014 ]; multiple scientists interacting with visualizations using multi-touch tables [Forlines and Lilien, 2008 ; Logtenberg, 2008 ; Logtenberg, 2009 ]; using Kinect and/or Leap Motion, targeted at scientists and learners [Alsayegh, Paramonov and Makatsoris, 2013 ; Hsiao, Cooper et al., 2014 ; Hsiao, Sun et al., 2016 ; Jamie and McRae, 2011 ; Sabir et al., 2013 ]; camera control with a separate touchscreen device [Lam and Siu, 2017 ]; as well as tangible [Sankaranarayanan et al., 2003 ] and “mixed reality” models [Gillet et al., 2005 ].
Overall, previous work in multi-touch molecular manipulation has largely focused on experts, use of simplified energy models or only visualizations, and often only one type of manipulation. In this work, we focus on the situation most relevant to a citizen science use case of multi-touch: users with a variety of scientific backgrounds, a complex energy function, and multiple types of manipulation.
3 Game design
In this work, we implemented a multi-touch interface for three common interactions in Foldit. Foldit already provided mouse interactions for these three tasks. A summary of the interactions used is given in Table 1 . First, Foldit allows the player to change the camera orientation, position and zoom. It is necessary to have different views onto the protein in order to be able to explore and manipulate the structure. Second, Foldit implements a pull action in order to flexibly change the protein structure itself. The protein structure changes so that the selected part of the protein follows the position of the pull action. Last, players have to be able to change the position of a protein in order to position different protein parts relative to each other.
Mouse camera. The camera can be changed by a left click and drag on the background in order to change the camera rotation around two axes. This is done by projecting the previous and current mouse positions onto a sphere in the center of the viewport. The axis and angle of the rotation between the two projected points are calculated and used to rotate the camera. The camera can also be translated using a right click and drag on the background. This is implemented similar to the rotation. The previous and current mouse positions are projected into the world space and the intersections with the plane in the center of the viewport are calculated. The translation between the two points on the plane is then applied to the camera. The camera zoom can be changed using the scroll wheel of the mouse. The relative changes of the mouse wheel are applied to a zoom factor, moving the camera closer to or further away from the center of the viewport.
Mouse pull. The protein structure can be changed flexibly using the pull action. This action can be initiated by a left click on part of the protein followed by a drag in the desired direction. An arrow between the selected part of the protein and the current mouse position indicates the pull action and visualizes the direction of the protein manipulation. The underlying software library Rosetta that is used for the computational modeling and analysis of the protein structures in Foldit recalculates the structure of the protein. This is done by locking the part of the protein which is furthest away from the point of interaction in place and adding a constraint that the selected part of the protein is as close as possible to a projected world space point of the current mouse position. The protein structure is constantly recalculated during the action and changes flexibly and dynamically. The action ends by releasing the pressed left mouse button.
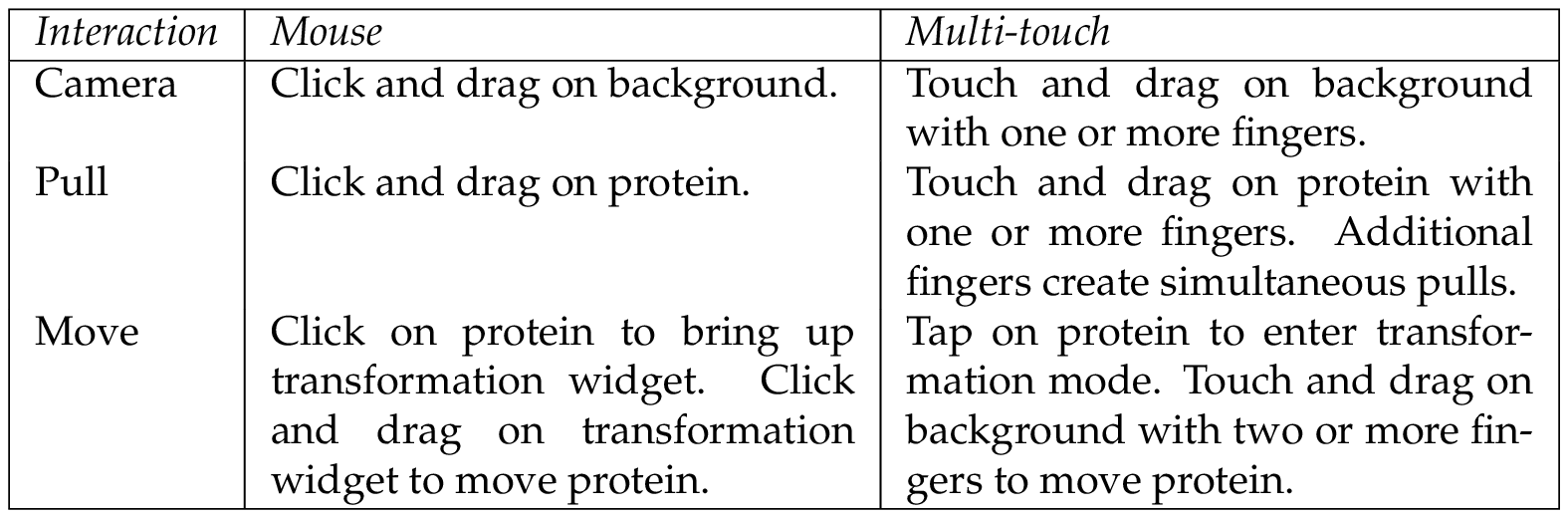
Mouse move. In some cases, proteins consist of multiple individual parts that have to be positioned in a specific relationship to each other in order to form a low energy composition. For this case, it is necessary to change the position of the protein parts relative to each other. Translation and rotation actions can be initiated by clicking once onto the protein with the left mouse button. A transformation widget consisting of two crossed arrows and a third curved arrow appears on release of the mouse, shown in Figure 1 . Left clicking on the arrows and dragging rotates the protein around the center of the transformation widget. Rotation around three axes is available to the user by dragging the mouse left/right, up/down and by dragging on the curved arrow in order to rotate along the view axis of the camera. Using a right click and drag on the transformation widget performs a translation. The translation is dependent on the view direction of the camera. The protein can be translated left/right and up/down on a plane in world space, keeping the distance to the camera.
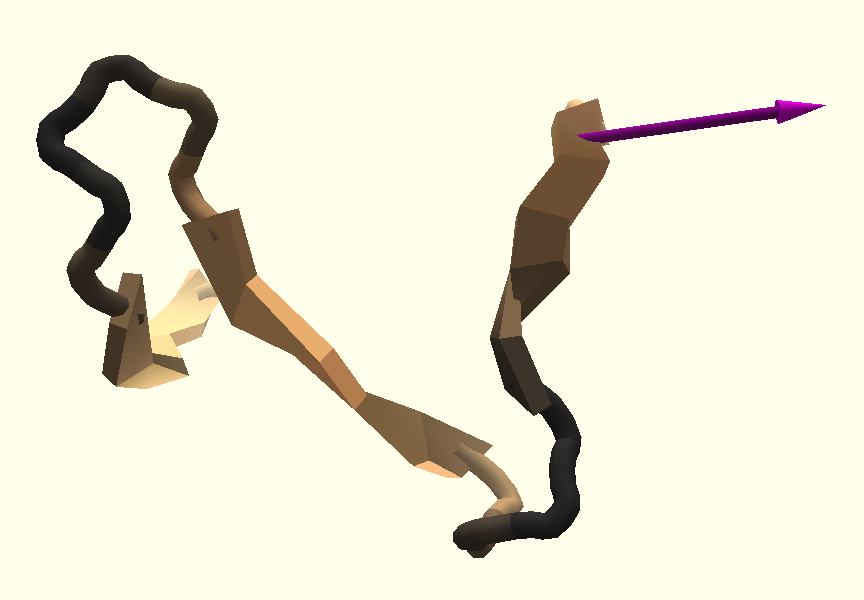
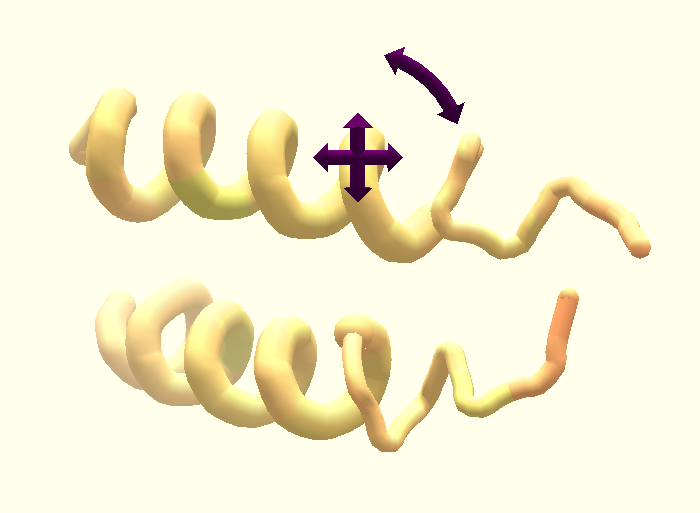
3.1 Multi-touch
For this work we implemented a multi-touch interface, making it possible to play Foldit on a touchscreen. The implemented touch gestures are based on best practice interface guidelines 1 2 and on touch interface guidelines from Wigdor and Wixon [Wigdor and Wixon, 2011 ]. In order to be able to interact with the standard game interface, e.g. select action, undo, etc., we implemented multi-touch such that if only one touch point is active it simulates a mouse event, making it possible to interact with game in a familiar fashion. If more than one touch point is active, they are used to implement touch-based transformations, e.g. pinch and zoom.
Multi-touch camera. The camera can be changed in 6 DOFs in total — rotation around three axes and translation in 3D space. With one finger touch and drag on the background the rotation around an axis can be controlled in a similar way as with the mouse. Additional controls can be performed with two or more fingers. When touching the background and moving with two or more fingers we calculate a 2D transformation between the previous and current finger positions, using a least squares estimator for non-reflective similarity transformation matrices. Such transformations are affine transformations with translation, rotation, and/or uniform scaling, and without reflection or shearing [Palén, 2016 ]. The translation is projected onto a plane in the center of the viewport in order to form a 3D transformation in world space, which is then applied to the camera. The 2D rotation of the transformation is applied along the view axis of the camera rotating the camera in 3D space. The scale calculated by the transformation is directly applied to the zoom factor of the camera moving it closer or further away from the center of the viewport. This technique allows the user to use familiar gestures like translate, rotate and zoom with two or more fingers to operate the camera.
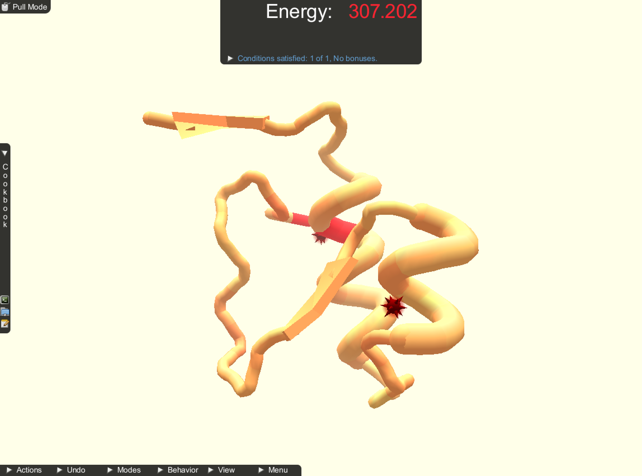
Multi-touch pull. While the mouse-based interface is only capable of pull actions at a single point of the protein, the multi-touch interface allows for pull interactions at multiple points at the same time. In order to empower players to manipulate proteins more according to their intention, we extended the interface to allow for multiple pull actions at the same time, shown in Figure 2 . Each touch on a protein starts a new pull action at this point of the protein. While dragging, the constrained recalculation of the protein structure receives constraints to best match each protein point to each touch point. While using the mouse, the protein was locked in place at the furthest part away from the part that was pulled. This is no longer feasible for multiple pull actions at the same time. Therefore, we unlock the position of the protein if more than one pull is performed at the same time and lock it again when pull actions are finished.

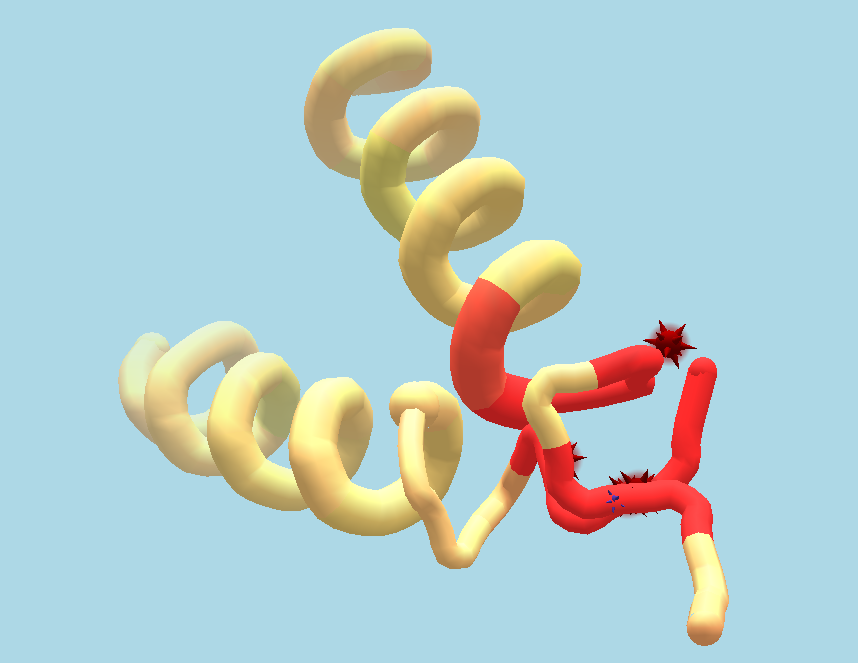
Multi-touch move. Individual protein parts can be controlled using multi-touch gestures similar to the camera controls. Because protein structures can have fine detailed structures and can occupy very little screen space it could be hard to perform protein transformations by touching the protein itself with multiple fingers. In addition, the protein structure would be covered by the fingers, making it harder to see and bring to the desired position or orientation. In order to avoid these problems, we implemented a dedicated transformation mode, which is activated by tapping onto the protein once. The transformation mode is activated for the selected protein part and is indicated by a change of color in the background from beige to light blue, seen in Figure 2 . In the transformation mode, two or more finger gestures on the background now transform the protein instead of the camera. The same technique used for calculating the transformation for the camera is used but now applied to the protein. The camera orientation can still be controlled using one finger drag in this mode. A second tap on the protein is used to leave the transformation mode.
4 Study design
We carried out a within-subjects experiment comparing the mouse and multi-touch Foldit interfaces. Each interface had interactions designed for two types of manipulations described above: pull , which allows the user to flexibly manipulate the protein backbone, and move , which allows the user to perform rigid body transformations. Additionally, users could manipulate the camera.
During manipulations, the current energy of the structure was shown at the top of the screen. Subjects could undo or redo to any point in their history of manipulations using a graph of energy history. All protein structures were made up of only glycine to simplify interactions, forgoing the need for sidechain manipulation, optimization and packing. Some tasks additionally contained a transparent guide visualization giving subjects a hint as to where to place the protein. This was meant to assist subjects, who may not have biochemistry experience, and thus may not fully understand the energy function, showing where to place the protein, but leaving it up to them how to get it there. The study as it is described here is approved by institutional review board at Wellesley College.
4.1 Procedure
At the beginning of the study, each subject was assigned to use one of the two interfaces to try to find the lowest energy they could. This is an open-ended task, which requires some understanding of the underlying energy function and time to explore the space of possible solutions; it would potentially be quite challenging for a novice.
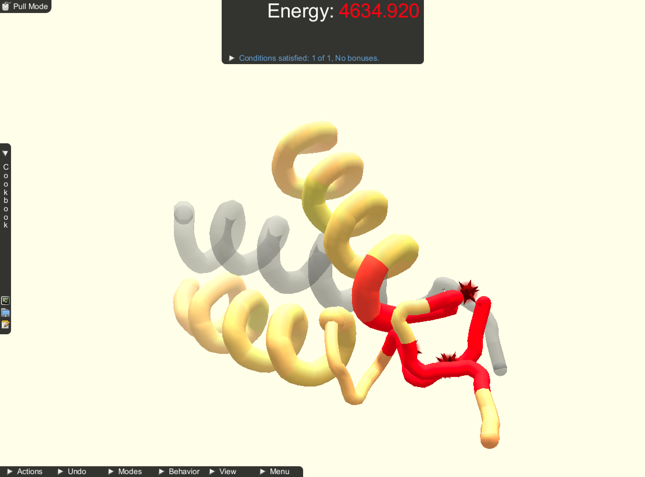
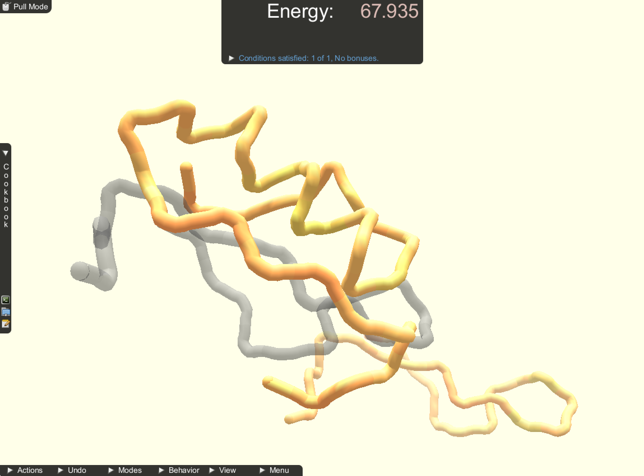
Subjects were given a short training overview of how to perform the two manipulations and camera control using the provided interface. Subjects were able to practice until they felt comfortable before moving on to the tasks. Subjects were also given a reference sheet of the interactions available. Subjects were then asked to complete two docking and two folding tasks. The docking tasks were set up with two protein parts, focused on move manipulations, and had guides. The folding tasks were set up with one protein part, focused on pull manipulations, and did not have guides. The tasks were always given in the same order, and subjects could use either manipulation in each. A screenshot of each fold task is shown in Figure 3 and of each docking task in Figure 4 . During these tasks, subject interactions were recorded. Subjects were prompted to move on if they were still working on a task after 5 minutes.
After completing all tasks for an interface, subjects were given a questionnaire about the interface consisting of questions from the System Usability Scale (SUS) [Brooke, 1996 ], the NASA Task Load Index (NASA-TLX) [Hart and Staveland, 1988 ], and the 4-item scales for Attention Allocation and Spatial Presence: Self-Location from the MEC Spatial Presence Questionnaire (MEC-SPQ) [Vorderer et al., 2004 ]. Once participants had completed the interface questionnaire for the first interface, the procedure for the training overview and experimental tasks were repeated for the second interface. Subjects then responded to the interface questionnaire for the second interface.
After completing both interfaces, subjects selected which of the two interfaces they preferred, and were given the option to do a “free play” session, for up to 15 minutes, consisting of three additional optional tasks. At the conclusion, subjects completed a short questionnaire consisting of questions about their demographics and overall experience.
20 participants began the study with the mouse interface and 21 participants began with the multi-touch interface. Subjects were assigned the interface used first to counterbalance gender and previous game and domain experience. The experiment was run on a Dell Inspiron laptop with a 15” touchscreen with a three-button mouse. The entire study took approximately one hour.
4.2 Participants
Subjects were recruited from students at Northeastern University and Wellesley College. 42 subjects were recruited, with one excluded due to a procedural error, leaving 41 subjects — 21 males and 20 females. 10 participants were classified as domain novices (taken one biochemistry class at the college level) and 9 participants were classified as domain experts (taken multiple biochemistry classes or work as a biochemist). 5 subjects self-identified as novice players of the game, and 1 self-identified as an expert player. All other subjects had never played Foldit before the study.
5 Results
During tasks, subjects’ actions and energies were logged, and in dock tasks, the alpha carbon root mean squared distance (RMSD) from the guide was also logged. For each participant, we summed the two values for each type of task to compute:
- Best Energy: the best (lowest) energy found during the task.
- Time to Best Energy: the time (in seconds) taken to find the best energy.
- Camera Count: the number of times the camera was adjusted.
- Pull Count: the number of pull manipulations.
- Move Count: the number of move manipulations.
- Undo Count: the number of times undo was used.
- Last RMSD (only for dock tasks): the RMSD to the guide at the end of the task.
These metrics were gathered in order to analyze and compare player performance between the two interfaces. We recorded Best Energy and Time to Best Energy representing high score and required time as standard performance measures in games. Camera, Pull and Move counts were measured in order to compare the number of moves need to accomplish the task. The difference in the interface might have an effect on the number of moves required to solve Foldit puzzles. We measured the Undo count as an indicator for mistakes performed by the players. In the docking tasks, we additionally measured RMSD as a measure for precision of the two interfaces.
A summary for fold tasks is given in Table 2 and for dock tasks in Table 3 . Based on paired-sample t-tests, only three comparisons were significant. In fold tasks, use of the multi-touch interface resulted in significantly more camera adjustments. In dock tasks, use of the multi-touch interface resulted in significantly fewer uses of the move manipulation and significantly lower ending RMSD.
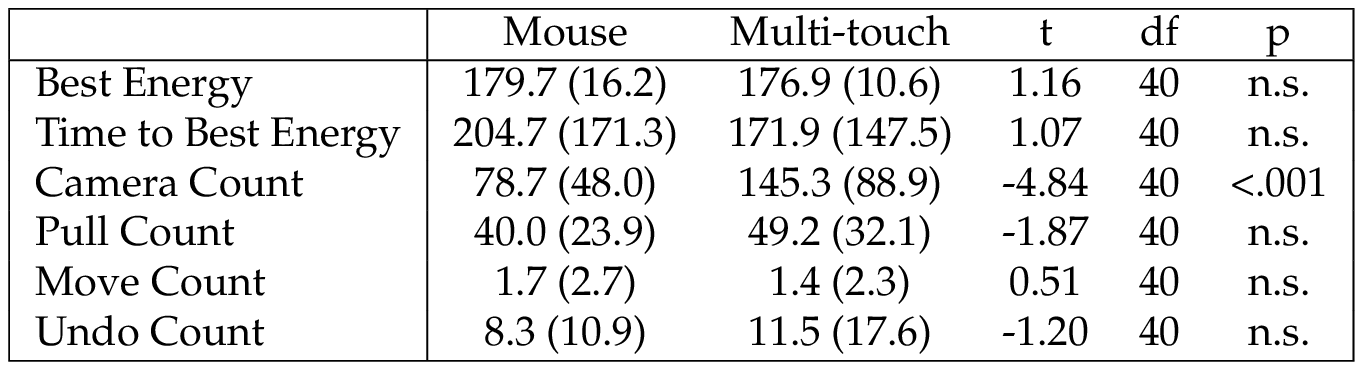
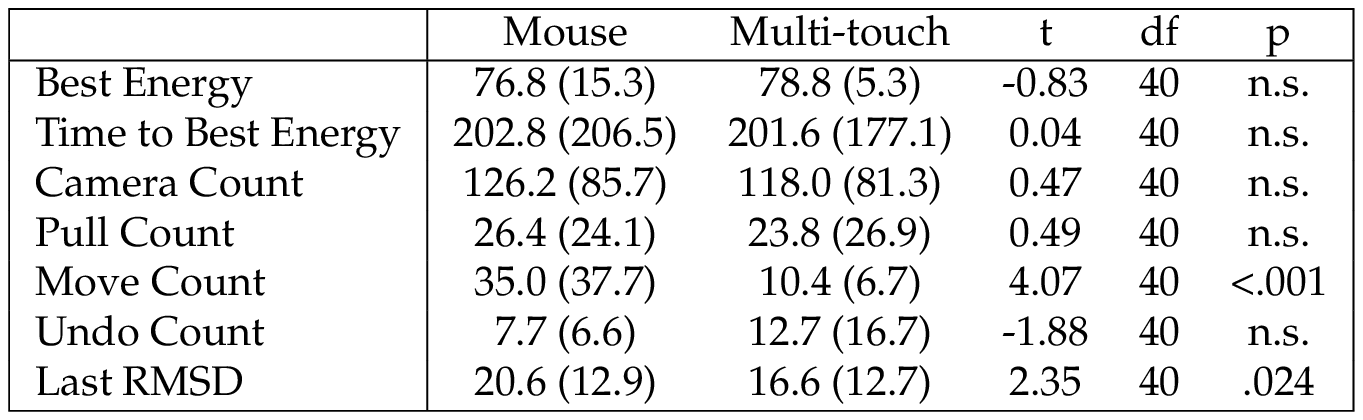
Table 4 summarizes the results from the interface questionnaires. Based on paired-sample t-tests, there were no significant differences for any of the unweighted NASA-TLX scores, enjoyment scores, or usability scores between the mouse and multi-touch interfaces. However, users reported significantly higher attention allocation and self-located spatial presence after using the multi-touch interface, compared to after using the mouse. For the “free play” session, 20 subjects selected mouse and 21 selected multi-touch, and a binomial test found no significant difference.
6 Discussion
Overall, subject performance was very similar in the two interfaces. Neither outperformed the other in terms of the best energy that was found or the time taken to find it. These results indicate that Foldit can potentially be played on touch screens without compromises in usability and player performance, compared to the traditional mouse based interface. Our study could show that this is true for manipulation of glycine structures with a subset of the tools that are normally available in the Foldit game. We argue that with a suitable interaction design, other tools could be properly implemented with a touch interface, as touch interaction offers a wide range of manipulation techniques. For this study, we decided to evaluate the interface only with simplified glycine structures, as this focused on the types of manipulations we examined, which dealt with the protein backbone. Further studies with advanced Foldit players are necessary to demonstrate that our findings are also true for more complex protein structures and additional tools for manipulation. In more practical real-world cases, for example, players might use multi-touch to manipulate the protein backbone, while the sidechains are automatically packed or optimized during the manipulation.
The fact that few of the participants had played Foldit before, compared to the large number who had never played the game, could have had an impact on the result of this study. We designed the study to be suitable for all types of participants as described above. However, further work could evaluate the impact of prior experience in Foldit on interface performance and preference. It may be interesting to explore if Foldit experts can better apply one or the other of the interfaces. In addition, previous research on Foldit by Cooper et al. [Cooper et al., 2010 ] found that the game is approachable by a wide variety of people, not only those with a biochemistry or scientific background.
In contrast to much prior work, we found largely similar performance and experience between the two interfaces. The result of equal performance in mouse and touch based interfaces is similar to the findings of Besançon et al. [Besançon et al., 2017 ], comparing mouse, touch, and tangible input modalities. However, in our work, the similar performance for both interfaces may have been impacted by the challenge of understanding the relatively complex energy function in the short training period. That is, for new Foldit players, it may be more of a challenge to decide what change should be made to the structure, than to make that change by manipulating the structure. When provided with an explicit guide in the dock tasks, subjects were able to better match the guide with the multi-touch interface, improving RMSD by nearly 20%, and able to do so using fewer than a third of the move manipulations, on average. In the case of fold tasks, the only difference was in the number of camera adjustments — with subjects in the multi-touch interface using nearly twice as many as in the mouse interface. While speculative, it may be the case this is due to the subject’s finger and hand occluding the protein while they were using pull manipulations (as occlusions are a common issue with touch-based selection [Forlines, Wigdor et al., 2007 ; Cockburn, Ahlström and Gutwin, 2012 ] ). This may have required the subjects to manipulate the camera more often to get a better perspective on the protein. As the multi-touch move manipulation was performed by touching the background rather than the protein, it may not have been as susceptible to such occlusions.
Surprisingly, neither mouse nor multi-touch interface was preferred by more users — there was an almost perfectly even split. This is consistent with the results from our subjective measures for enjoyment/frustration and usability, showing no significant differences. The significant differences in perceived attention and spatial presence may be caused by the directness of the multi-touch interface. Secondary effects of improved attention for the multi-touch interface, like higher scores or faster completion times were not found in our study, but may be present in a study looking at the mid- and long-term learning effects in the Foldit game. This would be an interesting question for future work as understanding the complex energy function of proteins in such a short time available in our study can be difficult. Further development of the touch interface for Foldit could support more of the protein manipulation functions provided by the game.
7 Conclusion
We have developed a multi-touch interface for the citizen science game Foldit and evaluated it in a comparative user study with 41 subjects against the standard mouse based interface. While subjects performed similarly overall regardless of interface, the results indicate that for tasks involving guided rigid body manipulations to dock protein parts, the multi-touch interface may allow players to complete tasks more accurately in fewer moves.
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant Nos. CNS-1513077 and CNS-1629879, and the German Academic Exchange Service by the program: BremenIDEA out.
References
-
Alsayegh, R., Paramonov, L. and Makatsoris, C. (2013). ‘A novel virtual environment for molecular system design’. In: 2013 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA) , pp. 37–42. https://doi.org/10.1109/civemsa.2013.6617392 .
-
Besançon, L., Issartel, P., Ammi, M. and Isenberg, T. (2017). ‘Mouse, tactile and tangible input for 3D manipulation’. In: Proceedings of the 2017 CHI Conference on Human Factors in Computing Systems — CHI ’17 . ACM, pp. 4727–4740. https://doi.org/10.1145/3025453.3025863 .
-
Brooke, J. (1996). ‘SUS — a quick and dirty usability scale’. Usability evaluation in industry 189 (194), pp. 4–7.
-
Cockburn, A., Ahlström, D. and Gutwin, C. (2012). ‘Understanding performance in touch selections: tap, drag and radial pointing drag with finger, stylus and mouse’. International Journal of Human-Computer Studies 70 (3), pp. 218–233. https://doi.org/10.1016/j.ijhcs.2011.11.002 .
-
Cohé, A., Dècle, F. and Hachet, M. (2011). ‘tBox: a 3d transformation widget designed for touch-screens’. In: Proceedings of the 2011 annual conference on Human factors in computing systems — CHI ’11 . New York, U.S.A.: ACM, pp. 3005–3008. https://doi.org/10.1145/1978942.1979387 .
-
Cooper, S., Khatib, F., Treuille, A., Barbero, J., Lee, J., Beenen, M., Leaver-Fay, A., Baker, D., Popović, Z. and Players, F. (2010). ‘Predicting protein structures with a multiplayer online game’. Nature 466 (7307), pp. 756–760. https://doi.org/10.1038/nature09304 .
-
Deterding, S., Dixon, D., Khaled, R. and Nacke, L. (2011). ‘From game design elements to gamefulness: defining “gamification”’. In: Proceedings of the 15th International Academic MindTrek Conference on Envisioning Future Media Environments — MindTrek ’11 (Tampere, Finland). New York, U.S.A.: ACM Press, pp. 9–15. https://doi.org/10.1145/2181037.2181040 .
-
Drucker, S. M., Fisher, D., Sadana, R., Herron, J. and Schraefel, M. C. (2013). ‘TouchViz: a case study comparing two interfaces for data analytics on tablets’. In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems — CHI ’13 . New York, U.S.A.: ACM, pp. 2301–2310. https://doi.org/10.1145/2470654.2481318 .
-
Fiorella, D., Sanna, A. and Lamberti, F. (2010). ‘Multi-touch user interface evaluation for 3D object manipulation on mobile devices’. Journal on Multimodal User Interfaces 4 (1), pp. 3–10. https://doi.org/10.1007/s12193-009-0034-4 .
-
Forlines, C. and Lilien, R. (2008). ‘Adapting a single-user, single-display molecular visualization application for use in a multi-user, multi-display environment’. In: Proceedings of the working conference on Advanced visual interfaces — AVI ’08 . New York, U.S.A.: ACM, pp. 367–371. https://doi.org/10.1145/1385569.1385635 .
-
Forlines, C., Wigdor, D., Shen, C. and Balakrishnan, R. (2007). ‘Direct-touch vs. mouse input for tabletop displays’. In: Proceedings of the SIGCHI conference on Human factors in computing systems — CHI ’07 . New York, U.S.A.: ACM, pp. 647–656. https://doi.org/10.1145/1240624.1240726 .
-
Gillet, A., Sanner, M., Stoffler, D. and Olson, A. (2005). ‘Tangible interfaces for structural molecular biology’. Structure 13 (3), pp. 483–491. https://doi.org/10.1016/j.str.2005.01.009 .
-
Hancock, M. S., Carpendale, S., Vernier, F. D., Wigdor, D. and Shen, C. (2006). ‘Rotation and translation mechanisms for tabletop interaction’. In: First IEEE International Workshop on Horizontal Interactive Human-Computer Systems (TABLETOP ’06) . IEEE, p. 8. https://doi.org/10.1109/tabletop.2006.26 .
-
Hancock, M., Carpendale, S. and Cockburn, A. (2007). ‘Shallow-depth 3d interaction: design and evaluation of one-, two-and three-touch techniques’. In: Proceedings of the SIGCHI conference on Human factors in computing systems — CHI ’07 . ACM, pp. 1147–1156. https://doi.org/10.1145/1240624.1240798 .
-
Hancock, M., Ten Cate, T. and Carpendale, S. (2009). ‘Sticky tools: full 6DOF force-based interaction for multi-touch tables’. In: Proceedings of the ACM International Conference on Interactive Tabletops and Surfaces — ITS ’09 . ACM, pp. 133–140. https://doi.org/10.1145/1731903.1731930 .
-
Hart, S. G. and Staveland, L. E. (1988). ‘Development of NASA-TLX (Task Load Index): results of empirical and theoretical research’. In: Human mental workload. Ed. by P. A. Hancock and N. Meshkati. Vol. 52. Amsterdam, The Netherlands: North Holland, pp. 139–183. https://doi.org/10.1016/s0166-4115(08)62386-9 .
-
Herrlich, M., Walther-Franks, B. and Malaka, R. (2011). ‘Integrated rotation and translation for 3D manipulation on multi-touch interactive surfaces’. In: Smart Graphics. Springer, pp. 146–154. https://doi.org/10.1007/978-3-642-22571-0_16 .
-
Hoggan, E., Nacenta, M., Kristensson, P. O., Williamson, J., Oulasvirta, A. and Lehtiö, A. (2013). ‘Multi-touch pinch gestures: performance and ergonomics’. In: Proceedings of the 2013 ACM international conference on Interactive tabletops and surfaces — ITS ’13 . New York, U.S.A.: ACM. https://doi.org/10.1145/2512349.2512817 .
-
Hoggan, E., Williamson, J., Oulasvirta, A., Nacenta, M., Kristensson, P. O. and Lehtiö, A. (2013). ‘Multi-touch rotation gestures: performance and ergonomics’. In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems — CHI ’13 . New York, U.S.A.: ACM. https://doi.org/10.1145/2470654.2481423 .
-
Hsiao, D.-Y., Cooper, S., Ballweber, C. and Popović, Z. (2014). ‘User behavior transformation through dynamic input mappings’. In: Proceedings of the 9th International Conference on the Foundations of Digital Games .
-
Hsiao, D.-Y., Sun, M., Ballweber, C., Cooper, S. and Popović, Z. (2016). ‘Proactive sensing for improving hand pose estimation’. In: Proceedings of the 2016 CHI Conference on Human Factors in Computing Systems — CHI ’16 . New York, U.S.A.: ACM. https://doi.org/10.1145/2858036.2858587 .
-
Jamie, I. M. and McRae, C. R. (2011). ‘Manipulating molecules: using Kinect for immersive learning in chemistry’. In: Proceedings of The Australian Conference on Science and Mathematics Education (formerly UniServe Science Conference) . Vol. 17.
-
Jankowski, J. and Hachet, M. (2015). ‘Advances in interaction with 3D environments’. Computer Graphics Forum 34 (1), pp. 152–190. https://doi.org/10.1111/cgf.12466 .
-
Kim, J. S., Greene, M. J., Zlateski, A., Lee, K., Richardson, M., Turaga, S. C., Purcaro, M., Balkam, M., Robinson, A., Behabadi, B. F., Campos, M., Denk, W. and Seung, H. S. (2014). ‘Space-time wiring specificity supports direction selectivity in the retina’. Nature 509 (7500), pp. 331–336. https://doi.org/10.1038/nature13240 .
-
Knoedel, S. and Hachet, M. (2011). ‘Multi-touch RST in 2D and 3D spaces: studying the impact of directness on user performance’. In: Proceedings of the 2011 IEEE Symposium on 3D User Interfaces (3DUI ’11) (Washington, DC, U.S.A.). IEEE, pp. 75–78. https://doi.org/10.1109/3dui.2011.5759220 .
-
Lam, W. W. T. and Siu, S. W. I. (2017). ‘PyMOL mControl: manipulating molecular visualization with mobile devices’. Biochemistry and Molecular Biology Education 45 (1), pp. 76–83. https://doi.org/10.1002/bmb.20987 .
-
Leaver-Fay, A., Tyka, M., Lewis, S. M., Lange, O. F., Thompson, J., Jacak, R., Kaufman, K. W., Renfrew, P. D., Smith, C. A., Sheffler, W., Davis, I. W., Cooper, S., Treuille, A., Mandell, D. J., Richter, F., Ban, Y.-E. A., Fleishman, S. J., Corn, J. E., Kim, D. E., Lyskov, S., Berrondo, M., Mentzer, S., Popović, Z., Havranek, J. J., Karanicolas, J., Das, R., Meiler, J., Kortemme, T., Gray, J. J., Kuhlman, B., Baker, D. and Bradley, P. (2011). ‘Rosetta3: an object-oriented software suite for the simulation and design of macromolecules’. Methods in Enzymology 487, pp. 545–574. https://doi.org/10.1016/b978-0-12-381270-4.00019-6 .
-
Levinthal, C. (1966). ‘Molecular model-building by computer’. Scientific American 214 (6), pp. 42–52. https://doi.org/10.1038/scientificamerican0666-42 .
-
Liu, J., Au, O. K.-C., Fu, H. and Tai, C.-L. (2012). ‘Two-finger gestures for 6DOF manipulation of 3D objects’. Computer Graphics Forum 31 (7), pp. 2047–2055. https://doi.org/10.1111/j.1467-8659.2012.03197.x .
-
Logtenberg, J. (2008). Designing interaction with molecule visualizations on a multi-touch table . Research for Capita Selecta, a Twente University course. Enschede, The Netherlands: Twente University.
-
— (2009). ‘Multi-user interaction with molecular visualizations on a multi-touch table’. MSc thesis. Enschede, The Netherlands: Twente University.
-
Ming, O.-Y., Beard, D. V. and Brooks, F. P. (1989). ‘Force display performs better than visual display in a simple 6D docking task’. In: Proceedings of the 1989 International Conference on Robotics and Automation . Vol. 3, pp. 1462–1466. https://doi.org/10.1109/robot.1989.100185 .
-
Palén, A. (2016). ‘Advanced algorithms for manipulating 2d objects on touch screens’. MSc thesis. Tampere, Finland: Tampere University of Technology.
-
Reisman, J. L., Davidson, P. L. and Han, J. Y. (2009). ‘A screen-space formulation for 2D and 3D direct manipulation’. In: Proceedings of the 22nd annual ACM symposium on User interface software and technology — UIST ’09 . New York, U.S.A.: ACM, pp. 69–78. https://doi.org/10.1145/1622176.1622190 .
-
Rousset, É., Bérard, F. and Ortega, M. (2014). ‘Two-finger 3D rotations for novice users: surjective and integral interactions’. In: Proceedings of the 2014 International Working Conference on Advanced Visual Interfaces — AVI ’14 . ACM, pp. 217–224. https://doi.org/10.1145/2598153.2598183 .
-
Sabir, K., Stolte, C., Tabor, B. and O’Donoghue, S. I. (2013). ‘The molecular control toolkit: controlling 3D molecular graphics via gesture and voice’. In: 2013 IEEE Symposium on Biological Data Visualization (BioVis) , pp. 49–56. https://doi.org/10.1109/biovis.2013.6664346 .
-
Sankaranarayanan, G., Weghorst, S., Sanner, M., Gillet, A. and Olson, A. (2003). ‘Role of haptics in teaching structural molecular biology’. In: 11th Symposium on Haptic Interfaces for Virtual Environment and Teleoperator Systems, 2003. HAPTICS 2003. Proceedings , pp. 363–366. https://doi.org/10.1109/haptic.2003.1191312 .
-
Surles, M. C., Richardson, J. S., Richardson, D. C. and Brooks, F. P. (1994). ‘Sculpting proteins interactively: continual energy minimization embedded in a graphical modeling system’. Protein Science 3 (2), pp. 198–210. https://doi.org/10.1002/pro.5560030205 .
-
Vorderer, P., Wirth, W., Gouveia, F. R., Biocca, F., Saari, T., Jäncke, F., Böcking, S., Schramm, H., Gysbers, A. and Hartmann, T. (2004). MEC spatial presence questionnaire (MEC-SPQ): short documentation and instructions for application . Report to the European Community, Project Presence: MEC (IST-2001-37661), 3.
-
Waldon, S. M., Thompson, P. M., Hahn, P. J. and Taylor, R. M. (2014). ‘SketchBio: a scientist’s 3D interface for molecular modeling and animation’. BMC Bioinformatics 15 (1), p. 334. https://doi.org/10.1186/1471-2105-15-334 .
-
Watson, D., Hancock, M., Mandryk, R. L. and Birk, M. (2013). ‘Deconstructing the touch experience’. In: Proceedings of the 2013 ACM international conference on Interactive tabletops and surfaces — ITS ’13 . New York, U.S.A.: ACM. https://doi.org/10.1145/2512349.2512819 .
-
Wigdor, D. and Wixon, D. (2011). Brave NUI world: designing natural user interfaces for touch and gesture. The Netherlands: Elsevier. https://doi.org/10.1016/B978-0-12-382231-4.00038-1 .
-
Wu, S., Chellali, A., Otmane, S. and Moreau, G. (2015). ‘TouchSketch: a touch-based interface for 3d object manipulation and editing’. In: Proceedings of the 21st ACM Symposium on Virtual Reality Software and Technology — VRST ’15 . New York, U.S.A.: ACM, pp. 59–68. https://doi.org/10.1145/2821592.2821606 .
Authors
Thomas Muender is a PhD Student at the Digital Media Lab of the University of Bremen. His research focuses on natural user interfaces for creating and manipulating 3D content in order to make it accessible for everyone. He graduated from University of Bremen with a master in computer science in 2016 and works at the lab of Rainer Malaka since then. E-mail: thom@uni-bremen.de .
Sadaab Ali Gulani is a computer science student at Northeastern University. He is working under Professor Seth Cooper on the citizen science game Foldit. E-mail: gulani.s@husky.neu.edu .
Lauren Westendorf worked as a Research Fellow and Lab Manager in the Wellesley Human Computer Interaction Lab from 2015–2017, where she focused on making complex data, such as synthetic biology and personal genomics, more approachable and understandable for non-experts and children. She graduated from Wellesley College in 2015, studying psychology and computer science. E-mail: lwestend@wellesley.edu .
Clarissa Verish is a research fellow at the Human Computer Interaction Lab at Wellesley College. She graduated from Wellesley in December 2016 with a degree in Chemistry. She is interested in engagement with science through tangible and digital interfaces. E-mail: cverish@wellesley.edu .
Rainer Malaka is Professor for Digital Media at the University of Bremen since 2006. The focus of Dr. Malaka’s work is Digital Media, Interaction, and Entertainment Computing. At the University of Bremen he directs a Graduate College “Empowering Digital Media” funded by the Klaus Tschira Foundation. He is also director of the center for computing technologies (TZI) at the University of Bremen. Before joining the University of Bremen, he lead a research group at the European Media Lab in Heidelberg. E-mail: malaka@tzi.de .
Orit Shaer is an Associate Professor of Computer Science and director of the Media Arts and Sciences Program at Wellesley College. She found and directs the Wellesley College Human-Computer Interaction (HCI) Lab. Her research focuses on next generation user interfaces including virtual and augmented reality, tangible and gestural interaction. She is a recipient of multiple NSF and industry awards including the prestigious NSF CAREER Award and Google App Engine Education Award. E-mail: oshaer@wellesley.edu .
Seth Cooper is an assistant professor in the College of Computer and Information Science at Northeastern University. He received his PhD in Computer Science and Engineering from the University of Washington. His current research focuses on using video games to solve difficult scientific problems; he is co-creator of the scientific discovery games Foldit and Nanocrafter and early math educational games including Refraction and Treefrog Treasure. E-mail: scooper@ccs.neu.edu .
Endnotes
1 Apple Gesture Guideline https://developer.apple.com/ios/human-interface-guidelines/user-interaction/gestures .
2 Android Gesture Guidelines https://material.io/guidelines/patterns/gestures.html .